Research
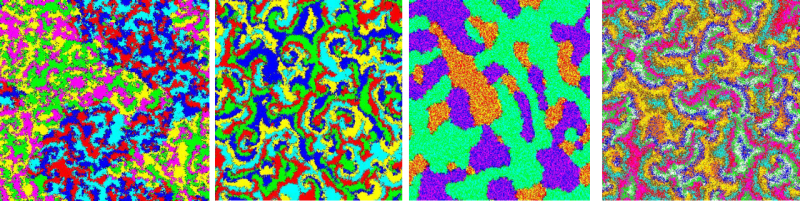
The research conducted at the Center for Soft Matter and Biological Physics covers a broad range of topics. Several key areas include:


Michel Pleimling
Professor
Department of Physics
Director of Academy of Integrated Science
Nonequilibrium Statistical Physics, Population Dynamics and Ecology, Soft/Bio Materials
Research Snapshots

Uwe Tauber
Professor
Department of Physics
Director of Center for Soft Matter and Biological Physics
Nonequilibrium Statistical Physics, Population Dynamics and Ecology, Cellular Biophysics, Soft/Bio Materials
Research Snapshots

Royce Zia
Professor emeritus
Department of Physics
Nonequilibrium Statistical Physics, Polymer Physics, Molecular Biophysics, Synthetic Biology

Justin Barone
Associate Professor
Department of Biological Systems Engineering
Polymer Physics, Soft/Bio Materials
Research Snapshots

Shengfeng Cheng
Assistant Professor
Department of Physics
Polymer Physics, Soft/Bio Materials, Molecular Biophysics
Research Snapshots

David Dillard
Professor
Department of Biomedical Engineering and Mechanics
Polymer Physics, Soft Matter Mechanics, Soft/Bio Materials

Herve Marand
Professor and Associate Chair
Department of Chemistry
Polymer Physics
Research Snapshots

Alexey Onufriev
Associate Professor
Department of Computer Science
Polymer Physics, Molecular Biophysics
Research Snapshots

Royce Zia
Professor emeritus
Department of Physics
Nonequilibrium Statistical Physics, Polymer Physics, Molecular Biophysics, Synthetic Biology

Louis Madsen
Associate Professor
Department of Chemistry
Soft/Bio Materials
Research Snapshots

David Dillard
Professor
Department of Biomedical Engineering and Mechanics
Polymer Physics, Soft Matter Mechanics, Soft/Bio Materials

Reza Mirzaeifar
Assistant Professor
Department of Mechanical Engineering
Soft Matter Mechanics, Soft/Bio Materials

C. Nadir Kaplan
Assistant Professor
Department of Physics
Soft Matter Mechanics, Soft/Bio Materials
Research Snapshots

Layne Watson
Professor
Department of Computer Sciences
Soft Matter Mechanics, Soft/Bio Materials, Cellular Biophysics

Justin Barone
Associate Professor
Department of Biological Systems Engineering
Polymer Physics, Soft/Bio Materials
Research Snapshots

Shengfeng Cheng
Assistant Professor
Department of Physics
Polymer Physics, Soft/Bio Materials, Molecular Biophysics
Research Snapshots

David Dillard
Professor
Department of Biomedical Engineering and Mechanics
Polymer Physics, Soft Matter Mechanics, Soft/Bio Materials

Louis Madsen
Associate Professor
Department of Chemistry
Soft/Bio Materials
Research Snapshots

Reza Mirzaeifar
Assistant Professor
Department of Mechanical Engineering
Soft Matter Mechanics, Soft/Bio Materials

C. Nadir Kaplan
Assistant Professor
Department of Physics
Soft Matter Mechanics, Soft/Bio Materials
Research Snapshots

Michel Pleimling
Professor
Department of Physics
Director of Academy of Integrated Science
Nonequilibrium Statistical Physics, Population Dynamics and Ecology, Soft/Bio Materials
Research Snapshots

Uwe Tauber
Professor
Department of Physics
Director of Center for Soft Matter and Biological Physics
Nonequilibrium Statistical Physics, Population Dynamics and Ecology, Cellular Biophysics, Soft/Bio Materials
Research Snapshots

Layne Watson
Professor
Department of Computer Sciences
Soft Matter Mechanics, Soft/Bio Materials, Cellular Biophysics

Daniel Capelluto
Associate Professor
Department of Biological Sciences
Molecular Biophysics
Research Snapshots

Shengfeng Cheng
Assistant Professor
Department of Physics
Polymer Physics, Soft/Bio Materials, Molecular Biophysics
Research Snapshots

Vinh Nguyen
Assistant Professor
Department of Physics
Molecular Biophysics

Alexey Onufriev
Associate Professor
Department of Computer Science
Polymer Physics, Molecular Biophysics
Research Snapshots

Royce Zia
Professor emeritus
Department of Physics
Nonequilibrium Statistical Physics, Polymer Physics, Molecular Biophysics, Synthetic Biology

Jing Chen
Assistant Professor
Department of Biological Sciences
Cellular Biophysics
Research Snapshots

Silke Hauf
Assistant Professor
Department of Biological Sciences
Cellular Biophysics

John Phillips
Professor
Department of Biological Sciences
Cellular Biophysics
Research Snapshots

Uwe Tauber
Professor
Department of Physics
Director of Center for Soft Matter and Biological Physics
Nonequilibrium Statistical Physics, Population Dynamics and Ecology, Cellular Biophysics, Soft/Bio Materials
Research Snapshots

Layne Watson
Professor
Department of Computer Sciences
Soft Matter Mechanics, Soft/Bio Materials, Cellular Biophysics

Royce Zia
Professor emeritus
Department of Physics
Nonequilibrium Statistical Physics, Polymer Physics, Molecular Biophysics, Synthetic Biology


Michel Pleimling
Professor
Department of Physics
Director of Academy of Integrated Science
Nonequilibrium Statistical Physics, Population Dynamics and Ecology, Soft/Bio Materials
Research Snapshots
Uwe Tauber
Professor
Department of Physics
Director of Center for Soft Matter and Biological Physics
Nonequilibrium Statistical Physics, Population Dynamics and Ecology, Cellular Biophysics, Soft/Bio Materials
Research Snapshots
Prof. Rana Ashkare

The Ashkar Group
Biological functions of cell membranes are controlled by the dynamic organization and interactions of the membrane's main building blocks, lipid and proteins. For simplicity, synthetic lipid membranes are usually studied as mimics of cell membranes in order to tease out reliable information of the interplay between membrane composition, structure, and dynamics and the resultant membrane functions. Using a suite of techniques including x-ray and neutron scattering and MD simulations, the aim of this project is to understand:
- Nanoscale membrane structure
- Effect of membrane composition on domain formation/structure and interactions
- Fast membrane dynamics and their role in biological functions
- Membrane bending and thickness fluctuations
- Membrane protein interactions
This project aims to investigate curvature effects in surface-modulated lipid membranes. This is done by utilizing the tunability of nanopatterned thermoresponsive polymer scaffolds to generate controllable 2D architectures in supported lipid membranes. The premise of this system is that it enables real-time realization of lateral membrane reorganization and peripheral protein binding in response to local membrane curvature, thus providing insights into critical membrane functions such as signal transduction, cell trafficking, and host-pathogen interactions. Further, such tunability of membrane topography can open new avenues to thermally switchable membrane-based biosensors.
Polymer nanocomposites (PNCs) are promising candidates for advanced multifunctional light-weight materials. The premise of PNCs lies in the myriad of possibilities they offer in synergistically integrating particle and polymer properties to obtain substantially improved material performance. Although earlier research on nanocomposites has remarkably enhanced our understanding of PNCs, less explored phenomena such as individual and collective chain dynamics and thermodynamically-driven wetting and dispersion phenomena, currently pose significant challenges to the applicability of PNCs in next-generation technologies. The focus of this project is to combine x-ray/neutron scattering with spectroscopy and simulations to understand the hierarchy of structures and dynamics and design advanced PNCs with controlled material properties.
Portraits of Virginia Tech College of Science faculty and staff

Applications of statistical physics to biological problems:
Glassy properties of prokaryotic bacteria; receptor-ligand binding kinetics
on cell membranes; predator-prey population dynamics -> movies;
cyclic competition models in ecology; evolutionary population dynamics
(mean-field and Smoluchowski theory; field theory; Monte Carlo simulations).
Dynamic critical behavior near equilibrium phase transitions:
Universality classes; anomalies in the ordered phase of isotropic systems;
crossover behavior; stability against non-equilibrium perturbations
(Langevin equations; dynamic field theory; renormalization group).
Phase transitions and scaling in systems far from equilibrium:
Directed percolation; Burgers/Kardar-Parisi-Zhang equation;
branching and annihilating random walks; diffusion-limited reactions;
driven diffusive systems; driven-dissipative Bose-Einstein condensation
(master and Langevin equations; field theory; renormalization group;
Monte Carlo simulations).
Statistical mechanics of flux lines in superconductors:
Mapping to boson quantum mechanics; influence of correlated disorder;
properties of the Bose glass phase; vortex transport and flux pinning;
critical properties of the normal- to superconducting transition with disorder;
voltage and flux density noise; non-equilibrium relaxation and aging features
(path integral description; Monte Carlo and Langevin dynamics simulations).
Structural phase transitions:
Influence of defects; dynamics; central peak
(Landau-Ginzburg theory of disordered systems; renormalization group).


